
doi: 10.1016/j.chroma.2004.03.022Įon-Duval A, Burke G (2004) Purification of pharmaceutical-grade plasmid DNA by anion-exchange chromatography in an RNase-free process. Teeters M, Root T, Lightfoot E (1036) Adsorption and desorption behavior of plasmid DNA on ion-exchange membranes: effect of salt valence and compaction agents. doi: 10.1016/j.chroma.2013.03.070Ĭhen WH, Fu JY, Kourentzi K, Willson RC (2011) Nucleic acid affinity of clustered-charge anion exchange adsorbents: effects of ionic strength and ligand density. Mota É, Sousa Â, Černigoj U, Queiroz JA, Tomaz CT, Sousa F (2013) Rapid quantification of supercoiled plasmid deoxyribonucleic acid using a monolithic ion exchanger. Tiainen P, Galaev I, Larsson PO (2007) Plasmid adsorption to anion-exchange matrices: comments on plasmid recovery. Sousa F, Queiroz JA (2011) Supercoiled plasmid quality assessment by analytical arginine-affinity chromatography. Ongkudon CM, Danquah MK (2010) Process optimisation for anion exchange monolithic chromatography of 4.2 kbp plasmid vaccine (pcDNA3F). Matos T, Queiroz JA, Bülow L (2013) Binding and elution behavior of small deoxyribonucleic acid fragments on a strong anion-exchanger multimodal chromatography resin. Yamamoto S, Nakamura M, Tarmann C, Jungbauer A (2007) Retention studies of DNA on anion-exchange monolith chromatography Binding site and elution behavior. Yamamoto S, Yoshimoto N, Tarmann C, Jungbauer A (2009) Binding site and elution behavior of DNA and other large biomolecules in monolithic anion-exchange chromatography. Ghanem A, Healey R, Adly FG (2013) Current trends in separation of plasmid DNA vaccines: a review. Liu MA (2011) DNA vaccines: an historical perspective and view to the future.


Pereira LR, Prazeres DMF, Mateus M (2010) Hydrophobic interaction membrane chromatography for plasmid DNA purification: design and optimization.
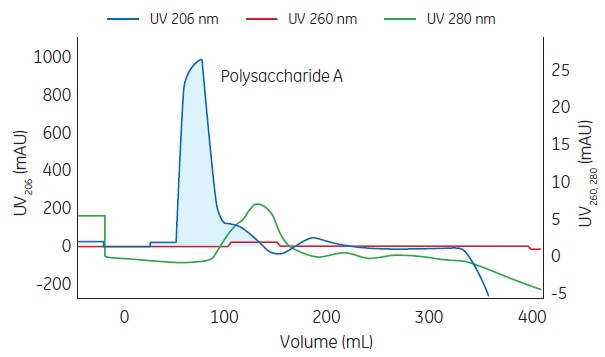
Sousa A, Sousa F, Queiroz JA (2012) Advances in chromatographic supports for pharmaceutical-grade plasmid DNA purification. This behaviour can be linked to the presence of the more hydrophobic phenyl group in Capto Adhere, leading to stronger retention of ssDNA molecules, which have a more hydrophobic character due to a higher degree of base exposure. Another pronounced difference between the resins was observed in the inverted elution of ss- and dsDNA, where ssDNA eluted at 2.88 M NaCl on Capto Adhere, while on Capto Q ImpRes ssDNA eluted already at 1.47 M NaCl. This recognition was not observed for Capto Adhere. Capto Q ImpRes provided a recognition for guanylate bases when samples of deoxynucleotides or poly(dG) were examined. All deoxynucleotides and DNAs tested bound strongly to the chromatographic materials and could be eluted by a linear gradient of increasing NaCl concentration. These variations in biophysical properties have been utilized for comparative separations on these two resins. The intrinsic differences between single- and double-stranded DNAs are related to charge, hydrophobicity, size and three-dimensional structure. Capto Adhere carries a multimodal ligand which combines strong anion with aromatic recognition, while Capto Q ImpRes is a strong anion exchanger with a chemically similar ligand, but without a phenyl group.

The differences in chromatographic behaviour of individual deoxynucleotides as well as small single-stranded and double-stranded DNA molecules have been examined for two resins from the Capto family: Capto Adhere and Capto Q ImpRes.


 0 kommentar(er)
0 kommentar(er)
